BLAST N Overview»
BLAST P Overview
SGI RASC Implementation
Documentation
FAQ
The BLASTN program consists of three stages, where the database being searched is streamed through a series of successive filters that have been configured by the search query. Each consecutive filter is more restrictive than the previous step. Patterns are either considered a match and passed on, or discarded in each filter. From analyzing and profiling the NCBI BLAST code, it is apparent that the vast majority of time is spent in the first two stages. As a result of these findings, Mitrionics' deployment focused on acceleration of the first two stages and left the third stage to the host processor.
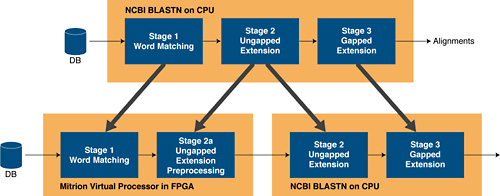
Figure 1. Three-stage deployment of BLAST.
In the FPGA-accelerated version of NCBI BLAST, the FPGA is loaded with a Mitrion Virtual Processor that has been programmed with, and adapted to, stage 1 and parts of stage 2 of the BLASTN program for nucleotide searches. The query is uploaded to the FPGA and the nucleotide database is streamed through. In stage one, the BLASTN program looks for exact patterns of a certain length (11 bases by default for nucleotide searches). The accelerated parts of stage 2 then process the matches, or "seeds," by extending them within a fixed window. The alignments that pass this stage are passed from the Mitrion Virtual Processor to the NCBI BLAST software on the host, where they are further extended, producing the final alignment sequences and the score based on the overall match.
Stage 1The first stage is divided into two sub-stages (see Figure 2 below). First a fast probabilistic filter returns a list of probable exact matches. This stage is implemented using bloom filters, a space-efficient probabilistic data structure that is used to test whether an element in the database is a member of the set of query words. Each bloom filter consists of a number of hash functions that check whether a specific bit is set or not in memory. This is where the FPGA architecture on the RC100 excels. Each FPGA has multiple independently accessible distributed memories that can all be used by the Mitrion Virtual Processor simultaneously each clock cycle. The flexibility of the Mitrion Virtual Processor allows these resources to be used where most needed and makes it possible to run a number bloom filters in parallel depending on the query size.
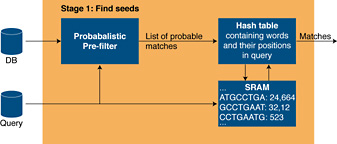
Figure 2. Stage 1 processing.
The bloom filters may produce false positives, but they will not produce any false negatives. The second part of stage 1 is a lookup in a hash table to filter out the false positives and to look up the position of the word within the query. Since the pre-filter discards the significant amounts of the initial data the performance requirements on the hash table lookup stage are much lower than on the bloom filters. The matches that pass the hash table look-up are forwarded to stage 2.
Stage 2The first part of the second stage extends the seed within a fixed window, returning matches that pass the threshold score and discard the rest. It is implemented as an unrolled systolic array; easily expressed as a Mitrion-C loop, which finds the best start position, stop position and score of two 64 character words in a single clock cycle.
All alignments that pass the threshold score are passed to a filter that discards matches that are so close to each other that they could not be extended without being joined. This part is done in stage 1 in the NCBI software implementation, but in the Mitrion implementation, it is executed after the ungapped extension part of stage 2 processing. This is due to the fact that it enhances the performance of the algorithm since the speed of the ungapped extension stage is so much better on the FPGA.
Matches that pass the final filter are returned from the FPGA to the host. Here, the un-accelerated parts of NCBI BLAST may extend them even further.
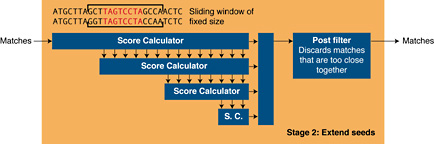
Figure 3. Stage 2 processing.
| All trademarks are the property of their owners. |